| Identification |
|---|
| Name | Monoglyceride lipase |
|---|
| Synonyms | - MGL
- Monoacylglycerol hydrolase
- Monoacylglycerol lipase
- Serine hydrolase YJU3
- MAG hydrolase
- MGH
- MAG lipase
- MAGL
|
|---|
| Gene Name | YJU3 |
|---|
| Enzyme Class | Not Available |
|---|
| Biological Properties |
|---|
| General Function | triglyceride metabolic process |
|---|
| Specific Function | Converts monoacylglycerides (MAG) to free fatty acids and glycerol. Required for efficient degradation of MAG, short-lived intermediates of glycerolipid metabolism which may also function as lipid signaling molecules. Controls inactivation of the signaling lipid N-palmitoylethanolamine (PEA). |
|---|
| Cellular Location | Not Available |
|---|
| SMPDB Pathways | | Triacylglycerol metabolism TG(12:0/14:0/25:0) | PW007788 | 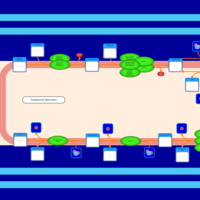   | | Triacylglycerol metabolism TG(12:0/14:0/26:0) | PW007800 | 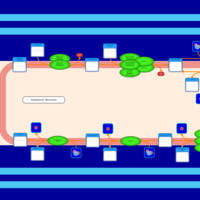   | | Triacylglycerol metabolism TG(12:0/14:0/27:0) | PW007816 |    | | Triacylglycerol metabolism TG(12:0/14:0/28:0) | PW007830 | 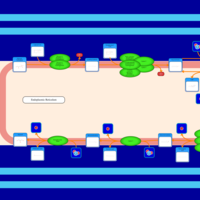 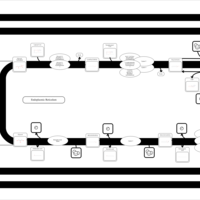 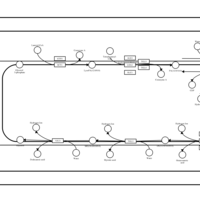 | | Triacylglycerol metabolism TG(12:0/14:0/29:0) | PW007848 | 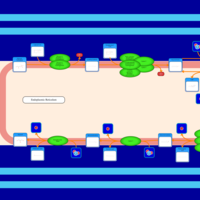 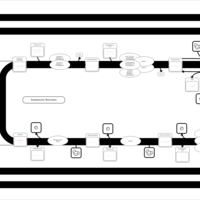 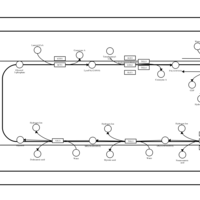 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions |
|
|---|
| KEGG Reactions | Not Available |
|---|
| Metabolites | | YMDB ID | Name | View |
|---|
| YMDB00069 | Palmitic acid | Show | | YMDB00283 | Glycerol | Show | | YMDB00862 | hydron | Show | | YMDB00890 | water | Show | | YMDB16219 | MG(14:1(9Z)/0:0/0:0) | Show | | YMDB16220 | MG(16:1(9Z)/0:0/0:0) | Show | | YMDB16221 | MG(18:1(9Z)/0:0/0:0) | Show | | YMDB16222 | MG(22:1(13Z)/0:0/0:0) | Show | | YMDB16223 | MG(24:1(15Z)/0:0/0:0) | Show |
|
|---|
| GO Classification | | Function |
|---|
| endoplasmic reticulum | | membrane | | lipid particle | | mitochondrial outer membrane | | acylglycerol lipase activity | | regulation of signal transduction | | triglyceride catabolic process | | triglyceride metabolic process |
|
|---|
| Gene Properties |
|---|
| Chromosome Location | Not Available |
|---|
| Locus | |
|---|
| Gene Sequence | >942 bp
ATGGCTCCGTATCCATACAAAGTGCAGACGACAGTACCTGAACTTCAATACGAAAACTTT
GATGGTGCTAAGTTCGGGTACATGTTCTGGCCTGTTCAAAATGGCACCAATGAGGTCAGA
GGTAGAGTTTTACTGATTCATGGGTTTGGCGAGTACACAAAGATTCAATTCCGGCTTATG
GACCACTTATCACTCAATGGTTACGAGTCATTTACGTTTGATCAAAGGGGTGCTGGTGTT
ACATCGCCGGGCAGATCGAAAGGTGTAACTGATGAGTACCATGTGTTTAACGATCTTGAG
CATTTTGTGGAGAAGAACTTGAGTGAATGTAAGGCCAAAGGCATACCCTTGTTCATGTGG
GGGCATTCAATGGGCGGTGGTATCTGCCTAAACTATGCCTGCCAAGGTAAGCACAAAAAC
GAAATAAGCGGATATATCGGGTCAGGCCCATTAATAATTTTACATCCGCATACAATGTAT
AACAAGCCGACCCAAATTATTGCTCCATTATTGGCGAAATTTTTACCAAGGGTAAGGATC
GACACTGGTTTAGATCTTAAAGGAATCACATCTGATAAAGCCTATCGTGCTTTCCTCGGA
AGCGATCCTATGTCTGTTCCACTATATGGGTCGTTTAGGCAAATACACGACTTTATGCAA
CGTGGTGCCAAGCTCTACAAGAATGAAAACAATTATATTCAGAAGAACTTCGCTAAAGAC
AAACCCGTTATTATTATGCATGGACAAGACGACACAATCAACGATCCTAAGGGCTCTGAA
AAGTTCATTCAGGACTGTCCTTCTGCTGACAAAGAATTAAAGCTGTATCCGGGCGCAAGA
CATTCGATTTTCTCACTAGAGACAGATAAAGTCTTCAACACGGTGTTCAATGATATGAAG
CAATGGTTGGACAAACACACCACGACCGAAGCTAAACCATAA |
|---|
| Protein Properties |
|---|
| Pfam Domain Function | Not Available |
|---|
| Protein Residues | 313 |
|---|
| Protein Molecular Weight | 35562.4 |
|---|
| Protein Theoretical pI | Not Available |
|---|
| PDB File | show |
|---|
| Signalling Regions | Not Available |
|---|
| Transmembrane Regions | Not Available |
|---|
| Protein Sequence | >Monoglyceride lipase
MAPYPYKVQTTVPELQYENFDGAKFGYMFWPVQNGTNEVRGRVLLIHGFGEYTKIQFRLM
DHLSLNGYESFTFDQRGAGVTSPGRSKGVTDEYHVFNDLEHFVEKNLSECKAKGIPLFMW
GHSMGGGICLNYACQGKHKNEISGYIGSGPLIILHPHTMYNKPTQIIAPLLAKFLPRVRI
DTGLDLKGITSDKAYRAFLGSDPMSVPLYGSFRQIHDFMQRGAKLYKNENNYIQKNFAKD
KPVIIMHGQDDTINDPKGSEKFIQDCPSADKELKLYPGARHSIFSLETDKVFNTVFNDMK
QWLDKHTTTEAKP |
|---|
| References |
|---|
| External Links | | Resource | Link |
|---|
| Saccharomyces Genome Database | YJU3 | | Uniprot ID | P28321 | | Uniprot Name | YJU3 | | PDB ID | 4ZWN |
|
|---|
| General Reference | Not Available |
|---|