| Identification |
|---|
| YMDB ID | YMDB02210 |
|---|
| Name | LysoPC(16:1(9Z)) |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Brewer's yeast |
|---|
| Description | LysoPC(16:1(9Z)) is a lysophospholipid (LyP). It is a monoglycerophospholipid in which a phosphorylcholine moiety occupies a glycerol substitution site. Lysophosphatidylcholines can have different combinations of fatty acids of varying lengths and saturation attached at the C-1 (sn-1) position. Fatty acids containing 16, 18 and 20 carbons are the most common. LysoPC(16:1(9Z)), in particular, consists of one chain of palmitoleic acid at the C-1 position. The palmitoleic acid moiety is derived from animal fats and vegetable oils. Lysophosphatidylcholine is found in small amounts in most tissues. It is formed by hydrolysis of phosphatidylcholine by the enzyme phospholipase A2, as part of the de-acylation/re-acylation cycle that controls its overall molecular species composition. It can also be formed inadvertently during extraction of lipids from tissues if the phospholipase is activated by careless handling. |
|---|
| Structure | |
|---|
| Synonyms | - 1-palmitoleoyl-glycero-3-phosphocholine
- LPC(16:1)
- LPC(16:1/0:0)
- LPC(16:1n7/0:0)
- LPC(16:1w7/0:0)
- LyPC(16:1)
- LyPC(16:1/0:0)
- LyPC(16:1n7/0:0)
- LysoPC(16:1)
- LysoPC(16:1/0:0)
- LysoPC(16:1n7/0:0)
- LysoPC(16:1w7/0:0)
- Lysophosphatidylcholine(16:1)
- Lysophosphatidylcholine(16:1/0:0)
- Lysophosphatidylcholine(16:1n7/0:0)
- Lysophosphatidylcholine(16:1w7/0:0)
- LyPC(16:1w7/0:0)
- 1-(9Z-Hexadecenoyl)-sn-glycero-3-phosphocholine
- 1-Palmitoleoyl-glycero-3-phosphocholine
- 1-Palmitoleoyl-glycerophosphocholine
- 1-Palmitoleoyl-GPC
- GPC(16:1(9Z))
- GPC(16:1)
- LPC 16:1(9Z)/0:0
- LPC(16:1(9Z)/0:0)
- LysoPC 16:1(9Z)/0:0
- Lysophosphatidylcholine(16:1(9Z)/0:0)
- PC 16:1(9Z)/0:0
- PC(16:1(9Z)/0:0)
- LPC(16:1)
- LPC(16:1/0:0)
- LPC(16:1n7/0:0)
- LPC(16:1W7/0:0)
- LyPC(16:1)
- LyPC(16:1/0:0)
- LyPC(16:1n7/0:0)
- LysoPC a C16:1
- LysoPC(16:1)
- LysoPC(16:1/0:0)
- LysoPC(16:1n7/0:0)
- LysoPC(16:1W7/0:0)
- Lysophosphatidylcholine(16:1)
- Lysophosphatidylcholine(16:1/0:0)
- Lysophosphatidylcholine(16:1n7/0:0)
- Lysophosphatidylcholine(16:1W7/0:0)
- LysoPC(16:1(9Z))
- 1-(9Z-Hexadecenoyl)-glycero-3-phosphocholine
- 1-Palmitoleoylglycerophosphocholine
- 1-Palmitoleoyl-lysophosphatidylcholine
- 1-Palmitoleoyl-sn-glycero-3-phosphocholine
- GPC(16:1(9Z)/0:0)
- GPC(16:1n7)
- GPC(16:1n7/0:0)
- GPC(16:1W7)
- GPC(16:1W7/0:0)
- LPC(16:1(9Z))
- LPC(16:1n7)
- LPC(16:1W7)
- LysoPC(16:1n7)
- LysoPC(16:1W7)
- Lysophosphatidylcholine(16:1(9Z))
- Lysophosphatidylcholine(16:1n7)
- Lysophosphatidylcholine(16:1W7)
- LysoPC(16:1(9Z)/0:0)
|
|---|
| CAS number | Not Available |
|---|
| Weight | Average: 493.6142
Monoisotopic: 493.316839407 |
|---|
| InChI Key | LFUDDCMNKWEORN-HSZRJFAPSA-N |
|---|
| InChI | InChI=1S/C24H48NO7P/c1-5-6-7-8-9-10-11-12-13-14-15-16-17-18-24(27)30-21-23(26)22-32-33(28,29)31-20-19-25(2,3)4/h10-11,23,26H,5-9,12-22H2,1-4H3/t23-/m1/s1 |
|---|
| IUPAC Name | (2-{[(2R)-3-[(9Z)-hexadec-9-enoyloxy]-2-hydroxypropyl phosphono]oxy}ethyl)trimethylazanium |
|---|
| Traditional IUPAC Name | (2-{[(2R)-3-[(9Z)-hexadec-9-enoyloxy]-2-hydroxypropyl phosphono]oxy}ethyl)trimethylazanium |
|---|
| Chemical Formula | C24H48NO7P |
|---|
| SMILES | CCCCCCC=CCCCCCCCC(=O)OC[C@@H](O)COP([O-])(=O)OCC[N+](C)(C)C |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1-acyl-sn-glycero-3-phosphocholines. These are glycerophosphocholines in which the glycerol is esterified with a fatty acid at O-1 position, and linked at position 3 to a phosphocholine. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphocholines |
|---|
| Direct Parent | 1-acyl-sn-glycero-3-phosphocholines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-acyl-sn-glycero-3-phosphocholine
- Phosphocholine
- Fatty acid ester
- Dialkyl phosphate
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- Tetraalkylammonium salt
- Quaternary ammonium salt
- Secondary alcohol
- Carboxylic acid ester
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Alcohol
- Organic oxygen compound
- Organopnictogen compound
- Carbonyl group
- Organic salt
- Amine
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | Not Available | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | | Lysolipid incorporation into ER PC(16:1(9Z)/16:1(9Z)) | PW002785 | 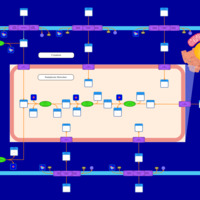 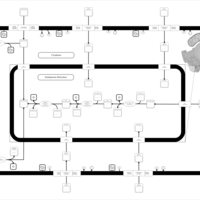 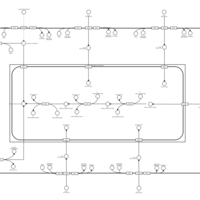 | | Lysolipid incorporation into Mitochondria PC(16:1(9Z)/16:1(9Z)) | PW002793 | 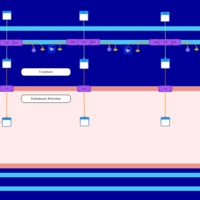 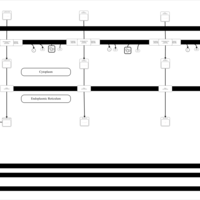 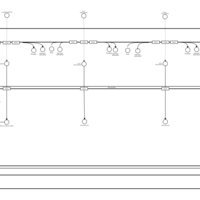 |
|
|---|
| KEGG Pathways | Not Available |
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | | Intracellular Concentration | Substrate | Growth Conditions | Strain | Citation |
|---|
| 0.443 ± 0.02215 µM | YEB media with 0.5 mM glucose | aerobic | Brewer's yeast | Experimentally Determined
Not Available | | 664000 ± 57825 umol/L | SD media with 2% raffinose | 24 oC | BY4741 | PMID: 19174513 | | 394250 ± 7875 umol/L | SD media with 2% raffinose | 37 oC | BY4741 | PMID: 19174513 | | Conversion Details Here |
|
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-009i-9421000000-8369cf40512bb91f9a92 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0000190000-3b2f823b3cd8feb26a69 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udo-0002990000-7176cdf7186de27b5716 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0j4l-0309300000-27e1bde7aced3e495eaf | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002f-0000900000-20d8b47f384d6c6abc9d | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001l-0900600000-d81e90dde6012dc43928 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0fai-0910400000-7b25c82c4a89b9527428 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0030900000-f418d5b39662ed44b8b0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-1090100000-eb1b8f27f35b0db065ee | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-1090000000-e8497356b3a95210d3f6 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000190000-d207603b0418f8cc497d | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ufr-0090070000-848973b728ef1ffc0d69 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-0090020000-121fe6e93b0d8bcfe171 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0000190000-fa3d2231e19f49a461ab | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0aor-0001960000-6bdc0b97b839b55840ee | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pcr-1609710000-c34b47c99b7f3709e023 | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|
