| Identification |
|---|
| YMDB ID | YMDB00913 |
|---|
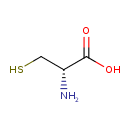
| Name | D-Cysteine |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | D-Cysteine, also known as D-cystein or DCY, belongs to the class of organic compounds known as cysteine and derivatives. Cysteine and derivatives are compounds containing cysteine or a derivative thereof resulting from reaction of cysteine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. D-Cysteine exists in all living species, ranging from bacteria to plants to humans. Based on a literature review a small amount of articles have been published on D-Cysteine. |
|---|
| Structure | |
|---|
| Synonyms | - D-Amino-3-mercaptopropionate
- D-Amino-3-mercaptopropionic acid
- (2S)-2-Amino-3-mercaptopropanoic acid
- (2S)-2-Amino-3-sulfanylpropanoic acid
- (S)-2-Amino-3-mercaptopropanoic acid
- D-Cystein
- D-Zystein
- DCY
- (2S)-2-Amino-3-mercaptopropanoate
- (2S)-2-Amino-3-sulfanylpropanoate
- (2S)-2-Amino-3-sulphanylpropanoate
- (2S)-2-Amino-3-sulphanylpropanoic acid
- (S)-2-Amino-3-mercaptopropanoate
- Cysteine
- Half cystine
- Cysteine hydrochloride
- Half-cystine
- L-Cysteine
- L Cysteine
- Zinc cysteinate
|
|---|
| CAS number | 921-01-7 |
|---|
| Weight | Average: 121.158
Monoisotopic: 121.019749163 |
|---|
| InChI Key | XUJNEKJLAYXESH-UWTATZPHSA-N |
|---|
| InChI | InChI=1S/C3H7NO2S/c4-2(1-7)3(5)6/h2,7H,1,4H2,(H,5,6)/t2-/m1/s1 |
|---|
| IUPAC Name | (2S)-2-amino-3-sulfanylpropanoic acid |
|---|
| Traditional IUPAC Name | L cysteine |
|---|
| Chemical Formula | C3H7NO2S |
|---|
| SMILES | [H]OC(=O)[C@]([H])(N([H])[H])C([H])([H])S[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cysteine and derivatives. Cysteine and derivatives are compounds containing cysteine or a derivative thereof resulting from reaction of cysteine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Cysteine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cysteine or derivatives
- Alpha-amino acid
- D-alpha-amino acid
- Amino acid
- Alkylthiol
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Primary amine
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Amine
- Hydrocarbon derivative
- Organopnictogen compound
- Organic oxide
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 220 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | 277 mg/mL at 25 oC [BEILSTEIN] | PhysProp | | LogP | -2.49 [HANSCH,C ET AL. (1995)] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | Not Available |
|---|
| KEGG Pathways | | Cysteine and methionine metabolism | ec00270 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004l-9100000000-4553906a941a5e87ec97 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-004i-9200000000-cfaf705cd0452d428454 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, N/A (Annotated) | splash10-056r-9300000000-ed3d829a54e4ac06325d | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, N/A (Annotated) | splash10-0a4i-9000000000-224425097c9b1883d266 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, N/A (Annotated) | splash10-0a4i-9000000000-39cb21d5fd8f893ba8fd | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00b9-9600000000-4352a7b437c34c04d9f0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-9200000000-7b4cd034c717561c61dd | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9000000000-8f351cbca6ffed356a9b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-6900000000-51ba0df80cc33423420b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0079-9400000000-4aa2b707c391d5838d29 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9000000000-95c7672c7836c0fc51c9 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00e9-8900000000-c05a34dee4bebd8457da | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9000000000-942ac689538269d6ca7b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9000000000-942ac689538269d6ca7b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a6r-9200000000-9d710aa92c54b66150d6 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-9000000000-e5d74af54cd557ea9df0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-f349eed712afbbc2670a | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Abe, T., Hashimoto, Y., Hosaka, H., Tomita-Yokotani, K., Kobayashi, M. (2008). "Discovery of amide (peptide) bond synthetic activity in Acyl-CoA synthetase." J Biol Chem 283:11312-11321.18305111
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|