| Identification |
|---|
| YMDB ID | YMDB00903 |
|---|
| Name | D-Proline |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | D-Proline, also known as D-prolin or DPR, belongs to the class of organic compounds known as proline and derivatives. Proline and derivatives are compounds containing proline or a derivative thereof resulting from reaction of proline at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. D-Proline is a very strong basic compound (based on its pKa). D-Proline exists in all living species, ranging from bacteria to humans. D-proline can be converted into 1-pyrroline-2-carboxylic acid through the action of the enzyme D-amino-acid oxidase. In yeast, D-proline is involved in the metabolic pathway called the arginine and proline metabolism pathway. D-Proline is a potentially toxic compound. |
|---|
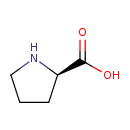
| Structure | |
|---|
| Synonyms | - (2R)-pyrrolidine-2-carboxylate
- (2R)-pyrrolidine-2-carboxylic acid
- (R)-pyrrolidine-2-carboxylate
- (R)-pyrrolidine-2-carboxylic acid
- D-Prolin
- D-proline
- R-Proline
- R)-2-Carboxypyrrolidine
- (R)-2-Carboxypyrrolidine
- DPR
- Proline
|
|---|
| CAS number | 344-25-2 |
|---|
| Weight | Average: 115.1305
Monoisotopic: 115.063328537 |
|---|
| InChI Key | ONIBWKKTOPOVIA-SCSAIBSYSA-N |
|---|
| InChI | InChI=1S/C5H9NO2/c7-5(8)4-2-1-3-6-4/h4,6H,1-3H2,(H,7,8)/t4-/m1/s1 |
|---|
| IUPAC Name | (2R)-pyrrolidine-2-carboxylic acid |
|---|
| Traditional IUPAC Name | D-proline |
|---|
| Chemical Formula | C5H9NO2 |
|---|
| SMILES | [H]OC(=O)[C@]1([H])N([H])C([H])([H])C([H])([H])C1([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as proline and derivatives. Proline and derivatives are compounds containing proline or a derivative thereof resulting from reaction of proline at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Proline and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Proline or derivatives
- Alpha-amino acid
- D-alpha-amino acid
- Pyrrolidine carboxylic acid
- Pyrrolidine carboxylic acid or derivatives
- Pyrrolidine
- Amino acid
- Carboxylic acid
- Secondary aliphatic amine
- Monocarboxylic acid or derivatives
- Secondary amine
- Organoheterocyclic compound
- Azacycle
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Amine
- Organopnictogen compound
- Organic oxide
- Carbonyl group
- Organic nitrogen compound
- Hydrocarbon derivative
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 221 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | 162 mg/mL at 25 oC [MERCK INDEX (1996)] | PhysProp | | LogP | -2.54 [HANSCH,C ET AL. (1995)] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | Not Available |
|---|
| KEGG Pathways | | Arginine and proline metabolism | ec00330 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00fu-9000000000-d203e3dfb0701403a75b | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-9000000000-56471980a375d26046ca | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9000000000-d3689ce7d1b666093d7f | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-00di-9200000000-0ff57624fa424cf13f20 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-03di-3900000000-01469d0f80dcbecd0185 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9000000000-b429bf31961274bc04e3 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0bt9-9600000000-5d761bf19a4ea84edc9c | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-6900000000-eba8ceb2a71d2726acd0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9200000000-c52871a4097a833a301a | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006x-9000000000-50a811fa0506a025048a | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-3900000000-ba371efbf747c7eed1a7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03k9-9600000000-f8a0815fbf4d1b2713af | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-133753f87329f1da7d39 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-3900000000-2d7e8539e5f8306d0afe | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ot-9400000000-b7d551a4cee7688ee082 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-a4e2ca2cba04978b1bf6 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-9000000000-97990d3a6f4d86f2e35e | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9000000000-6af75747a42ba65860f4 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9000000000-4d5b790a456b8349d65b | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- Horak, J., Rihova, L. (1982). "L-Proline transport in Saccharomyces cerevisiae." Biochim Biophys Acta 691:144-150.6753931
|
|---|
| Synthesis Reference: | Not Available |
|---|
| External Links: | |
|---|