| Identification |
|---|
| YMDB ID | YMDB00383 |
|---|
| Name | Gluconic acid |
|---|
| Species | Saccharomyces cerevisiae |
|---|
| Strain | Baker's yeast |
|---|
| Description | Gluconic acid, also known as D-gluconate or D-glukonsaeure, belongs to the class of organic compounds known as sugar acids and derivatives. Sugar acids and derivatives are compounds containing a saccharide unit which bears a carboxylic acid group. Gluconic acid is an extremely weak basic (essentially neutral) compound (based on its pKa). Gluconic acid exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | - 2-Dehydro-3-deoxy-D-gluconate
- 2-Keto-3-deoxy-D-gluconate
- 2,3,4,5,6-pentahydroxy-hexanoate
- 2,3,4,5,6-pentahydroxy-hexanoic acid
- 2,3,4,5,6-Pentahydroxyhexanoate
- 2,3,4,5,6-Pentahydroxyhexanoic acid
- D-gluco-hexonic acid
- D-gluconate
- D-gluconic acid
- D-gluconsaeure
- D-glukonsaeure
- Dextronate
- Dextronic acid
- GCO
- Glosanto
- Gluconate
- Gluconic acid
- Glycogenate
- Glycogenic acid
- Glyconate
- Glyconic acid
- Hexonic acid
- Magnesium D-gluconate dihydratae
- Magnesium gluconate
- Maltonate
- Maltonic acid
- Pentahydroxycaproate
- Pentahydroxycaproic acid
- Sodium stibogluconate
- (2R,3S,4R,5R)-2,3,4,5,6-Pentahydroxyhexanoic acid
- (2R,3S,4R,5R)-2,3,4,5,6-Pentahydroxyhexanoate
- D-Gluco-hexonate
- Hexonate
- Boron gluconate
- Gluconic acid, (113)indium-labeled
- Gluconic acid, calcium salt
- Gluconic acid, cesium(+3) salt
- Gluconic acid, lanthanum(+3) salt
- Gluconic acid, sodium salt
- Gluconic acid, strontium (2:1) salt
- Magnerot
- Manganese gluconate
- Sodium gluconate
- Zinc gluconate
- Gluconic acid, (159)dysprosium-labeled salt
- Gluconic acid, aluminum (3:1) salt
- Gluconic acid, ammonium salt
- Gluconic acid, magnesium (2:1) salt
- Gluconic acid, (14)C-labeled
- Gluconic acid, 1-(14)C-labeled
- Gluconic acid, 6-(14)C-labeled
- Gluconic acid, cobalt (2:1) salt
- Gluconic acid, copper salt
- Gluconic acid, manganese (2:1) salt
- Gluconic acid, potassium salt
- Gluconic acid, tin(+2) salt
- Gluconic acid, zinc salt
- Lithium gluconate
- Gluconic acid, (99)technecium (5+) salt
- Gluconic acid, fe(+2) salt, dihydrate
- Gluconic acid, monolithium salt
- Gluconic acid, monopotassium salt
- Gluconic acid, monosodium salt
|
|---|
| CAS number | 526-95-4 |
|---|
| Weight | Average: 196.1553
Monoisotopic: 196.058302738 |
|---|
| InChI Key | RGHNJXZEOKUKBD-SQOUGZDYSA-N |
|---|
| InChI | InChI=1S/C6H12O7/c7-1-2(8)3(9)4(10)5(11)6(12)13/h2-5,7-11H,1H2,(H,12,13)/t2-,3-,4+,5-/m1/s1 |
|---|
| IUPAC Name | (2R,3S,4R,5R)-2,3,4,5,6-pentahydroxyhexanoic acid |
|---|
| Traditional IUPAC Name | gluconate |
|---|
| Chemical Formula | C6H12O7 |
|---|
| SMILES | [H]OC(=O)[C@]([H])(O[H])[C@@]([H])(O[H])[C@]([H])(O[H])[C@]([H])(O[H])C([H])([H])O[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as sugar acids and derivatives. Sugar acids and derivatives are compounds containing a saccharide unit which bears a carboxylic acid group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Sugar acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Gluconic_acid
- Medium-chain hydroxy acid
- Medium-chain fatty acid
- Beta-hydroxy acid
- Hydroxy fatty acid
- Alpha-hydroxy acid
- Fatty acyl
- Fatty acid
- Hydroxy acid
- Monosaccharide
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Polyol
- Monocarboxylic acid or derivatives
- Alcohol
- Carbonyl group
- Primary alcohol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Charge | 0 |
|---|
| Melting point | 113-118 °C |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Water Solubility | 316 mg/mL at 25 oC [MERCK INDEX (1996)] | PhysProp | | LogP | Not Available | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Organoleptic Properties | Not Available |
|---|
| SMPDB Pathways | Not Available |
|---|
| KEGG Pathways | | Pentose and glucuronate interconversions | ec00040 | 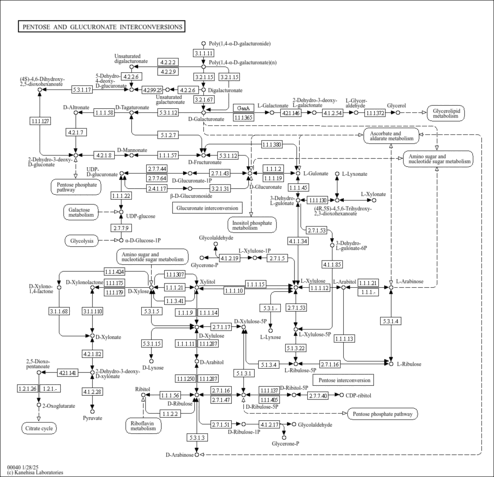 | | Pentose phosphate pathway | ec00030 |  |
|
|---|
| SMPDB Reactions | Not Available |
|---|
| KEGG Reactions | Not Available |
|---|
| Concentrations |
|---|
| Intracellular Concentrations | Not Available |
|---|
| Extracellular Concentrations | Not Available |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | View |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-0002-0932000000-202af87cea2d1f7184ab | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-005a-0920000000-2308d9356bc5bb01420e | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-014j-0950000000-61ab7adf15e353df4ba2 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (6 TMS) | splash10-0le9-1964000000-5eb7d6777170e1ad5fa0 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0002-0932000000-202af87cea2d1f7184ab | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0le9-1964000000-5eb7d6777170e1ad5fa0 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0002-0931000000-b07fcc4ac1e6701d32c8 | JSpectraViewer | MoNA | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0fr2-0930000000-181f7b697b591cf8080b | JSpectraViewer | MoNA | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-06tu-9500000000-ebe398c88f74bb51e702 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (6 TMS) - 70eV, Positive | splash10-03fr-6121297000-698854fa2453487a1d69 | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_6_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Gluconic acid,6TBDMS,#1" TMS) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | JSpectraViewer | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | JSpectraViewer | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-06vj-1900000000-78e4a5d92be678c068e4 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-03ds-5900000000-f5d8284baa473ce9d77f | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-014i-9500000000-7c5b20eef98d0e3d6cbb | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0900000000-b2632ca9154cc5e44438 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-002b-5900000000-4a3066f9dfd6653682fb | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-004i-9000000000-50f63dfd017a8380a47c | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-056r-9000000000-91ccf7c8949c1c9d852a | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0a6r-9000000000-6e40f1cebd8b460016b3 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-0900000000-b2632ca9154cc5e44438 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-002b-5900000000-cf9b480ac397acfebc71 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-9000000000-eca61f6a9c0f1d06e84d | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-056r-9000000000-91ccf7c8949c1c9d852a | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0a6r-9000000000-6e40f1cebd8b460016b3 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0a6r-9000000000-3950065b412843471434 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-0a6r-7900000000-5bb7335ada119405f6ad | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-056r-5900000000-717fb7f30be3f6d61d46 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-002b-6900000000-a8da6e6791f61d154360 | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-052b-9500000000-a346a36e3dd0bea4abcc | JSpectraViewer | MoNA | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-056r-9000000000-6e1dd3cd14f2b116fa7c | JSpectraViewer | MoNA | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004j-2900000000-42150530e3232b66059b | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-9500000000-763d7b03787732284c19 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bvi-9100000000-040569af699927ca6859 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-05ic-9800000000-3571d4f2ff79bc0d7fa0 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052r-9400000000-3cb207fadbe1fcae5cb7 | JSpectraViewer | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9100000000-8c900935a94065d0938b | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer | | 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available | JSpectraViewer | | 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |
|
|---|
| References |
|---|
| References: | - UniProt Consortium (2011). "Ongoing and future developments at the Universal Protein Resource." Nucleic Acids Res 39:D214-D219.21051339
- Scheer, M., Grote, A., Chang, A., Schomburg, I., Munaretto, C., Rother, M., Sohngen, C., Stelzer, M., Thiele, J., Schomburg, D. (2011). "BRENDA, the enzyme information system in 2011." Nucleic Acids Res 39:D670-D676.21062828
- SABLE, H. Z., GUARINO, A. J. (1952). "Phosphorylation of gluconate in yeast extracts." J Biol Chem 196:395-402.12980980
- Cadiere, A., Ortiz-Julien, A., Camarasa, C., Dequin, S. (2011). "Evolutionary engineered Saccharomyces cerevisiae wine yeast strains with increased in vivo flux through the pentose phosphate pathway." Metab Eng 13:263-271.21300171
- Castrillo, J. I., Zeef, L. A., Hoyle, D. C., Zhang, N., Hayes, A., Gardner, D. C., Cornell, M. J., Petty, J., Hakes, L., Wardleworth, L., Rash, B., Brown, M., Dunn, W. B., Broadhurst, D., O'Donoghue, K., Hester, S. S., Dunkley, T. P., Hart, S. R., Swainston, N., Li, P., Gaskell, S. J., Paton, N. W., Lilley, K. S., Kell, D. B., Oliver, S. G. (2007). "Growth control of the eukaryote cell: a systems biology study in yeast." J Biol 6:4.17439666
|
|---|
| Synthesis Reference: | Anastassiadis, Savas; Morgunov, Igor G. Gluconic acid production. Recent Patents on Biotechnology (2007), 1(2), 167-180. |
|---|
| External Links: | |
|---|


