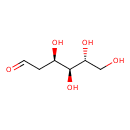
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 4 TMS) | splash10-0q29-1951000000-06cc16a9680628545167 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 4 TMS) | splash10-0lkc-0951000000-39e56efe41d6473040e4 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0fr5-0920000000-5f7dfef22a1b59cae284 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0ktb-0920000000-6f9638179f688d51022e | JSpectraViewer | MoNA |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ir0-9400000000-0d9dd630f3590782a74e | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-054t-9258700000-0738ee18a80ba8e5b90a | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-3900000000-45865b9816a45604d811 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01ow-9200000000-7b5a5cbd361ec11efbbf | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0707-9000000000-c9e6d3430eb1073f0d79 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0w29-5900000000-90300437f9ab1170e562 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0k96-9400000000-a11cb0518964ee7a83dd | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-b4574a9d519fced6b332 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-6900000000-319e473c192f4b110b8d | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9000000000-d161d9b754ebaeb29a74 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-0a62c56f2a4845f4d597 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-08ml-9700000000-9df4724be189ef01f655 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0abc-9000000000-e85603d6bed71b3882e0 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-4ed5058ebfd661cb8c61 | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |