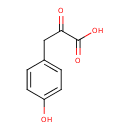
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-002f-1920000000-75b07d9c09371340939f | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-00xr-9340000000-087aad2497b3493d27c0 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 2 TMS) | splash10-002o-5910000000-fd1e55c84c79bfeca559 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 2 TMS) | splash10-002f-1941000000-2b4c87544657895fcaf6 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 3 TMS) | splash10-014i-3492100000-483bdcea11d60fe4d306 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 3 TMS) | splash10-014i-6791000000-ddc0a3b695cd1d0769f2 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-002f-1951000000-ecd071a64b26ca77684c | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-002f-1920000000-75b07d9c09371340939f | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00xr-9340000000-087aad2497b3493d27c0 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-002o-5910000000-fd1e55c84c79bfeca559 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-002f-1941000000-2b4c87544657895fcaf6 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-014i-3492100000-483bdcea11d60fe4d306 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-014i-6791000000-ddc0a3b695cd1d0769f2 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0006-2920000000-469469dfaad745fa78e8 | JSpectraViewer | MoNA |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-002o-2910000000-4f336335a2366e7dd6db | JSpectraViewer | MoNA |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-6900000000-91bfe3ee54a36877251b | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0adi-6961000000-670161f1542fda9ff1fe | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | JSpectraViewer |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | JSpectraViewer |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-0a4i-0900000000-12033042c41b550bed42 | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-0a4i-0900000000-6e8a3701c6d254cc824d | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0560-1900000000-8082c68e259d24234d05 | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-004i-0900000000-2416d7f64101b9473cbb | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0a4i-2900000000-b4c21b3d9751b9e56d67 | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0005-9400000000-708e258e692a0abfbc92 | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0002-9100000000-7d75e23ae6944c1e9c6d | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-001m-9000000000-fb80dcbab23323a042af | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-0900000000-2416d7f64101b9473cbb | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0a4i-2900000000-ad1f1fabdf6b1579fff7 | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0005-9400000000-708e258e692a0abfbc92 | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-9100000000-7d75e23ae6944c1e9c6d | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-001m-9000000000-fb80dcbab23323a042af | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-001i-1900000000-76ae33b45e4348d50b07 | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-02bf-7900000000-665940e67ad40f759048 | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-053i-9800000000-0f708ba46e75f553e77c | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-00dl-5900000000-86f6d19bba9a914d22ee | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-0159-0900000000-a1a6bf5b3a0ac3863c90 | JSpectraViewer | MoNA |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-06ur-9700000000-18795e8a2dc93ca36a93 | JSpectraViewer | MoNA |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-08gr-0900000000-be3cb913118057f499d1 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-06rj-0900000000-6bebea0fda2ef3a2263b | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-6900000000-ede11b81a97e891c66e3 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0900000000-2584f872c980915c1739 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01s9-1900000000-249c71cfa9cefe2a8020 | JSpectraViewer |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001l-3900000000-66991d861a91be9b9286 | JSpectraViewer |
| MS | Mass Spectrum (Electron Ionization) | splash10-0a59-5900000000-6c003e26816579df31ca | JSpectraViewer | MoNA |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 1H NMR Spectrum | Not Available | JSpectraViewer |
| 1D NMR | 13C NMR Spectrum | Not Available | JSpectraViewer |
| 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available | JSpectraViewer |
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | JSpectraViewer |